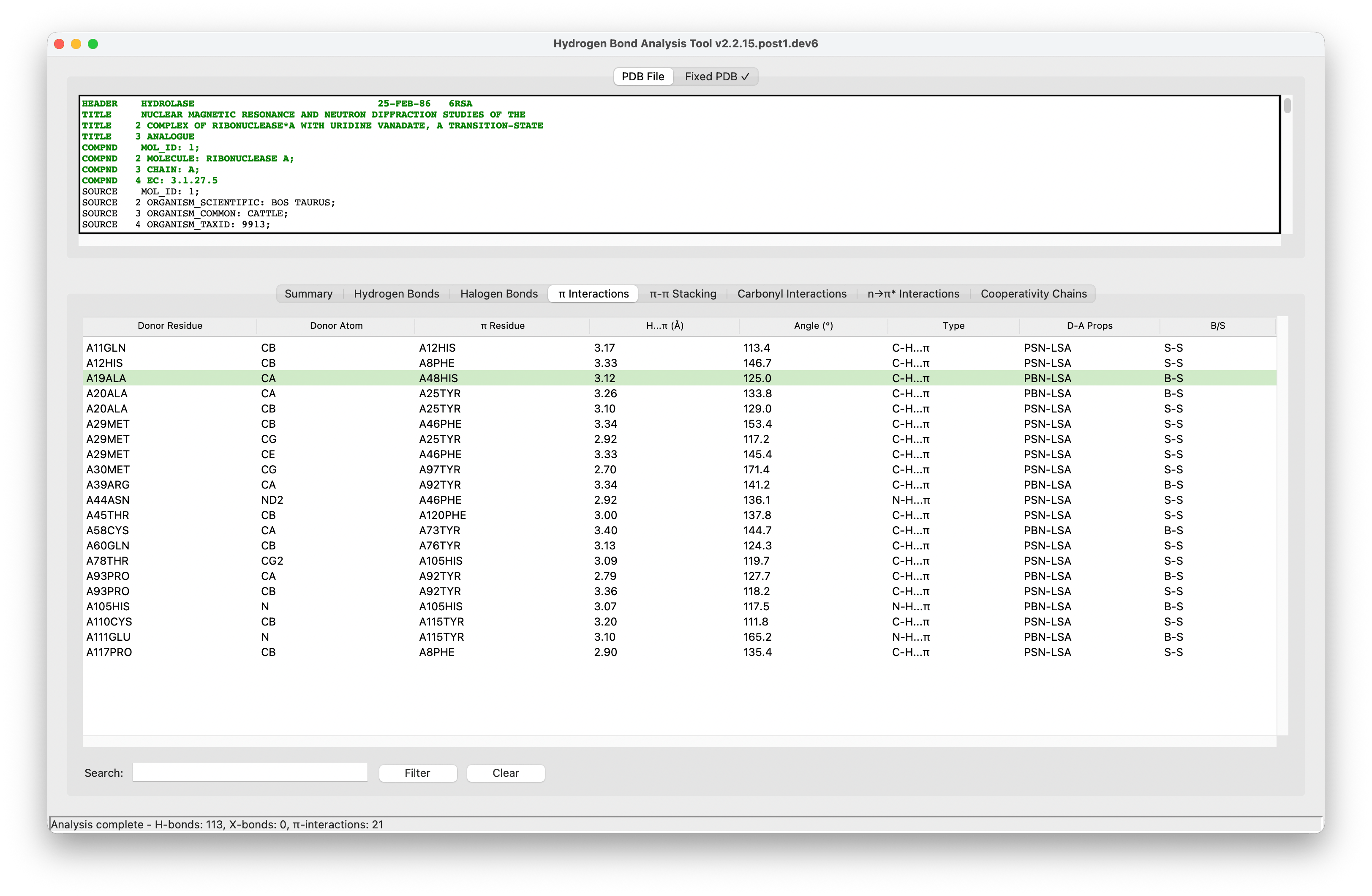
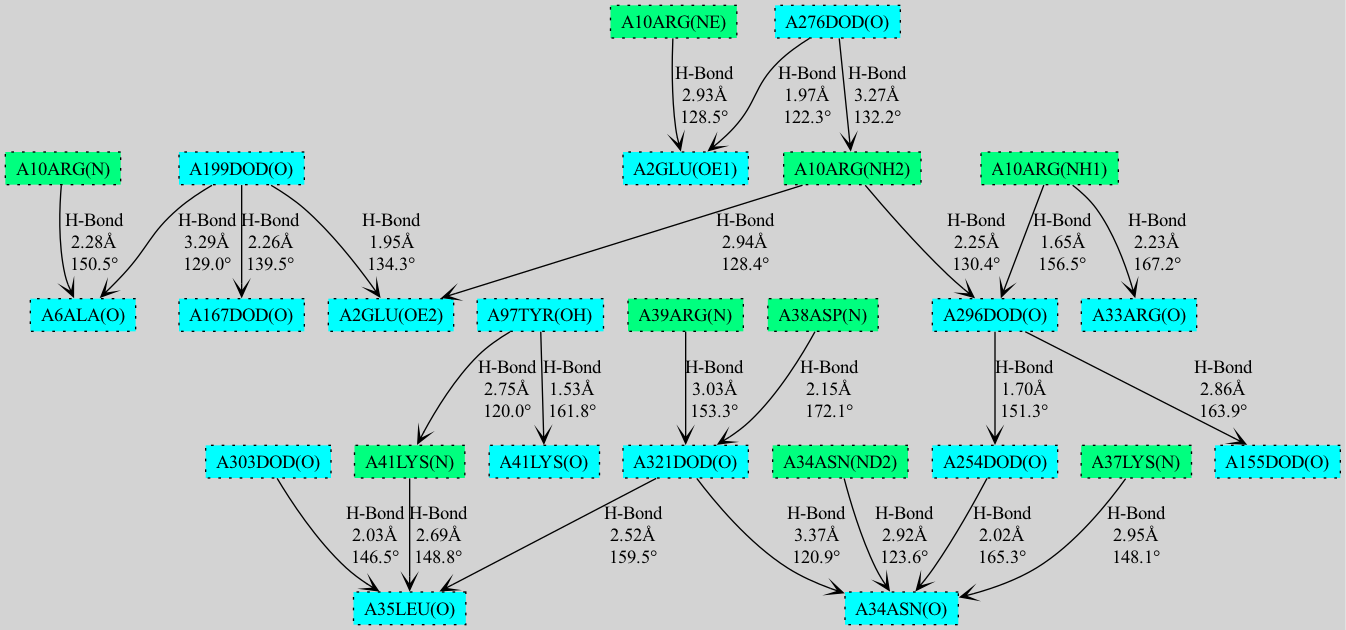
A Python package to automate the analysis of potential hydrogen bonds and similar type of weak interactions in macromolecular structures, available in Protein Data Bank (PDB) file format. HBAT2 uses a geometric approach to identify molecular interactions by analyzing distance and angular criteria.
Supported Interaction Types:
- Hydrogen Bonds: Classical
N-H···O,O-H···O, and weakC-H···Ointeractions - Halogen Bonds:
C-X···Ainteractions (X = Cl, Br, I) - π Interactions: X-H···π and
C-X···πinteractions with aromatic rings (Phe,Tyr,Trp,His, etc.) - π-π Stacking: Aromatic ring-ring interactions (parallel, T-shaped, offset)
- Carbonyl Interactions:
n→π*interactions between carbonyl groups - n-π Interactions: Lone pair interactions with aromatic
πsystems
HBAT2 is a modern Python re-implementation of the original Perl-based tool developed by Abhishek Tiwari and Sunil Kumar Panigrahi. HBAT v1 can still be downloaded from SourceForge however Perl version is not maintained anymore.
- Detect and analyze potential hydrogen bonds, halogen bonds, π interactions, π-π stacking, carbonyl interactions, and n-π interactions
- Automated PDB fixing with OpenBabel and PDBFixer integration
- Support graphical (tkinter), command-line, and programming API interfaces
- Use graphical interfaces for interactive analysis, CLI/API for batch processing and automation
- Cooperativity chain visualization using NetworkX/matplotlib and GraphViz
- Export cooperativity chain visualizations to PNG, SVG, PDF formats
- Built-in presets for different structure types (high-resolution, NMR, membrane proteins, etc.)
- Customizable distance cutoffs, angle thresholds, and analysis modes.
- Multiple Output Formats: Text, CSV, and JSON export options
- Optimized algorithms for efficient analysis of large structures
- Cross-Platform: Works on Windows, macOS, and Linux.
Please review HBAT documentation for more details.
pip install hbatRun HBAT Command-Line Interface (CLI) using hbat or launch HBAT GUI using hbat-gui.
git clone https://github.com/abhishektiwari/hbat.git
cd hbat
pip install -e .Alternatively,
pip install git+https://github.com/abhishektiwari/hbat.gitRun HBAT Command-Line Interface (CLI) using hbat or launch HBAT GUI using hbat-gui.
conda install -c hbat hbat
- Python: 3.9 or higher
- tkinter: tkinter is included with Python standard library on most systems. However, on Mac install Python and tkinter using
brew.
brew install python python3-tk
- GraphViz (Optional): Required for advanced cooperativity chain visualization with high-quality graph rendering. HBAT will automatically fall back to NetworkX/matplotlib visualization if GraphViz is not available.
Install GraphViz:
On Ubuntu/Debian:
sudo apt-get update
sudo apt-get install graphvizOn macOS (using Homebrew):
brew install graphvizOn Windows:
- Download and install from GraphViz official website
- Or using Chocolatey:
choco install graphviz - Or using conda:
conda install -c conda-forge graphviz
Note: After installing GraphViz, restart your terminal/command prompt before running HBAT to ensure the GraphViz executables are available in your PATH.
Launch the GUI application:
hbat-guiThe GUI provides,
- File browser for loading PDB files
- Parameter configuration panels
- Tabbed results display
- Export and visualization options
Basic usage:
hbat input.pdbHBAT supports multiple output formats:
# No output flag - displays results to console
hbat input.pdb # Display results to console
# Single file outputs (format auto-detected from extension)
hbat input.pdb -o results.txt # Text format (human-readable summary + details)
hbat input.pdb -o results.json # JSON format (single file with all interactions)
# Multiple file outputs (separate files per interaction type)
hbat input.pdb --csv results # Creates results_h_bonds.csv, results_x_bonds.csv, etc.
hbat input.pdb --json results # Creates results_h_bonds.json, results_x_bonds.json, etc.With custom parameters:
hbat input.pdb -o results.txt --hb-distance 3.0 --mode localhbat --list-presetshbat protein.pdb --preset high_resolution
hbat membrane_protein.pdb --preset membrane_proteinshbat protein.pdb --preset drug_design_strict --hb-distance 3.0 --verbosepositional arguments:
input Input PDB file
optional arguments:
-h, --help show this help message and exit
-o OUTPUT, --output OUTPUT
Output file (format auto-detected from extension: .txt, .json)
--json JSON Export to multiple JSON files (base name for files)
--csv CSV Export to multiple CSV files (base name for files)
Preset Options:
--preset PRESET Load parameters from preset file (.hbat or .json)
--list-presets List available example presets and exit
Analysis Parameters:
Hydrogen Bond Parameters:
--hb-distance HB_DISTANCE
Hydrogen bond H...A distance cutoff in Å (default: 2.5)
--hb-angle HB_ANGLE Hydrogen bond D-H...A angle cutoff in degrees (default: 120)
--da-distance DA_DISTANCE
Donor-acceptor distance cutoff in Å (default: 3.5)
Halogen Bond Parameters:
--xb-distance XB_DISTANCE
Halogen bond X...A distance cutoff in Å (default: 3.9)
--xb-angle XB_ANGLE Halogen bond C-X...A angle cutoff in degrees (default: 150)
π Interaction Parameters:
--pi-distance PI_DISTANCE
π interaction H...π distance cutoff in Å (default: 3.5)
--pi-angle PI_ANGLE π interaction D-H...π angle cutoff in degrees (default: 110)
π-π Stacking Parameters:
--pi-pi-distance PI_PI_DISTANCE
π-π centroid-to-centroid distance cutoff in Å (default: 3.8)
--pi-pi-parallel-angle PI_PI_PARALLEL_ANGLE
Maximum angle for parallel π-π stacking in degrees (default: 30.0)
--pi-pi-tshaped-angle-min PI_PI_TSHAPED_ANGLE_MIN
Minimum angle for T-shaped π-π stacking in degrees (default: 60.0)
--pi-pi-tshaped-angle-max PI_PI_TSHAPED_ANGLE_MAX
Maximum angle for T-shaped π-π stacking in degrees (default: 90.0)
--pi-pi-offset PI_PI_OFFSET
Maximum lateral offset for parallel π-π stacking in Å (default: 2.0)
Carbonyl Interaction Parameters (n→π*):
--carbonyl-distance CARBONYL_DISTANCE
Carbonyl O···C distance cutoff in Å (default: 3.2)
--carbonyl-angle-min CARBONYL_ANGLE_MIN
Minimum O···C=O angle for carbonyl interactions in degrees (default: 95.0)
--carbonyl-angle-max CARBONYL_ANGLE_MAX
Maximum O···C=O angle for carbonyl interactions in degrees (default: 125.0)
n→π* Interaction Parameters:
--n-pi-distance N_PI_DISTANCE
Lone pair to π center distance cutoff in Å (default: 3.6)
--n-pi-sulfur-distance N_PI_SULFUR_DISTANCE
Sulfur-specific distance cutoff in Å (default: 4.0)
--n-pi-angle-min N_PI_ANGLE_MIN
Minimum angle to π plane in degrees (default: 0.0)
--n-pi-angle-max N_PI_ANGLE_MAX
Maximum angle to π plane in degrees (default: 45.0)
General Parameters:
--covalent-factor COVALENT_FACTOR
Covalent bond detection factor (default: 0.85)
--mode {complete,local}
Analysis mode: complete (all interactions) or local (intra-residue only)
Output Control:
--verbose, -v Verbose output with detailed progress
--quiet, -q Quiet mode with minimal output
--summary-only Output summary statistics only
Analysis Filters:
--no-hydrogen-bonds Skip hydrogen bond analysis
--no-halogen-bonds Skip halogen bond analysis
--no-pi-interactions Skip π interaction analysis
--no-pi-pi-stacking Skip π-π stacking analysis
--no-carbonyl-interactions
Skip carbonyl n→π* interaction analysis
--no-n-pi-interactions
Skip n→π* interaction analysis
This project is licensed under the MIT License - see the LICENSE file for details.
If you use HBAT or HBAT2 in your research, please cite:
@article{tiwari2007hbat,
author = {Tiwari, Abhishek and Panigrahi, Sunil Kumar},
doi = {10.3233/ISI-2007-00337},
journal = {In Silico Biology},
month = dec,
number = {6},
title = {{HBAT: A Complete Package for Analysing Strong and Weak Hydrogen Bonds in Macromolecular Crystal Structures}},
volume = {7},
year = {2007}
}
@misc{tiwari_2025_17645321,
author = {Tiwari, Abhishek},
title = {HBAT2: A Python Package to analyse Hydrogen Bonds and Other Non-covalent Interactions in Macromolecular Structures},
month = nov,
year = 2025,
publisher = {Zenodo},
doi = {10.5281/zenodo.17645321},
url = {https://doi.org/10.5281/zenodo.17645321},
}
See our contributing guide and development guide. At a high-level,
- Fork the repository
- Create a feature branch
- Make your changes
- Add tests if applicable
- Submit a pull request